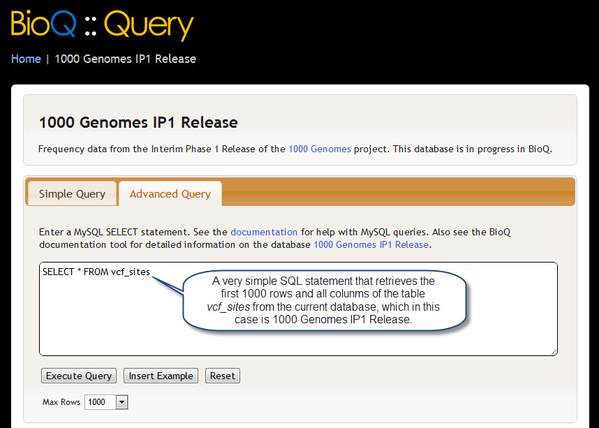
Advanced Queries with SQL
In the Advanced Query section of the BioQ::Query page you may enter a precise SQL query that will be passed to our MySQL database server. A tutorial on writing SQL queries can be found here, and general documentation on MySQL can be found here.
e
You can specify any table from the current database, which in thise case is 1000 Genomes IP1 Release. You can also access other databases using database.table notation, such as bioq_dbsnp_human_134.snp_summary.
The following special tables, which are determined from the Enter Genomic Features section, can also be used in an advanced query:
| Special Table | Description |
|---|---|
| my_snps | dbSNP SNP ID |
| my_vars | For 1000 Genomes queries only: variants identified by their genomic position using the pos_global identifier described here. |
| my_genes | Genes determined from the Enter Genomic Features section |
Note that dbSNP-Q will automatically insert a LIMIT statement into the query to limit the number rows retrieved. The default is 1000 rows, and can be changed via the Max Rows dropdown menu below the query editor. You may override this setting by using a LIMIT statement:
SELECT * FROM my_snps LIMIT 1000
There is an overall maximum number of rows that may be retrieved by any query regardless of the LIMIT statement - this is currently set to 10 million (to look this parameter up view the Program Parameters tab at the bottom of the query results section after executing a query and look for the parameter named limit-rows).
EXAMPLES OF ADVANCED QUERIES
The following examples use the dbSNP Build 134 database. Click the Execute query link below each example to execute the query in BioQ. Note that the query text may also be copied and pasted into the BioQ query editor, but be sure not to copy the line numbers.
A basic SELECT statement
The following query will retrieve all the columns for the first 20 rows of the table _loc_snp_summary. This is similar to theSNP Summary button in the Simple Query section but the custom query retrieves all the columns from _loc_snp_summary rather than a subset. Note that you may click on a row for a vertical view, which is useful when there are many columns.
SELECT * FROM _loc_snp_summary LIMIT 20
Note that this query has nothing to do with the SNPs entered in the Enter SNPs section. This type of query is useful for getting an idea of what kind of data exists in a table. To look up dbSNP information for the SNPs from the Enter SNPssection we must use a JOIN. This is discussed below.
Selecting specific columns
This query will select only the snp_id and ancestral_allele columns from the _loc_snp_summary table. Note that MySQL will ignore carriage returns, extra spaces, and so on.
SELECT snp_id, ancestral_alleles FROM _loc_snp_summary LIMIT 20
Selecting specific rows
The MySQL WHERE clause is used to select specific rows. Below are some examples of expressions that can be used with a WHERE clause. For more information see the MySQL online documentation.
Exact match
SELECT * FROM _loc_snp_summary WHERE (snp_id = 16969968)
Match a set of items
SELECT * FROM _loc_snp_summary WHERE (snp_id IN (16969968,1051730,4950))
Sorting the results of a query
Note that the results of any query in dbSNP-Q can be sorted by any single column by clicking on the label of that column in the results table. Sorting may also be done directly using the the MySQL ORDER BY clause which is useful if you wish to sort by multiple columns.
SELECT snp_id, rep_gene_symbol, unique_chr, unique_pos_bp
FROM _loc_snp_summary
WHERE (rep_gene_symbol IN ('CHRNA5','CHRNB3'))
ORDER BY unique_chr, unique_pos_bp
Note that in this example the table _loc_snp_summary stores the column unique_chr as character data and so the information is sorted treating the numbers as characters. For example "8" comes after "15".
Using SNPs from the Enter Genomic Features section in your queries
Some of the following examples require entries in the Enter Genomic Features section at the top of the BioQ query page. You can click Insert Example to insert example entries. Some of the Execute Query links below will automatically insert the example features.
The SNPs determined by the entries in the Enter Genomic Features section, whether they be entered directly by rs ID or derived from a gene or region query, are stored in a special table named my_snps. Use the following query to view the table my_snps. Note this table includes the column exists_in_dbsnp which is 1 or 0 depending on whether the SNP rs number, stored in the column snp_id, was found or not found in the dbSNP database, respectively.
SELECT * FROM my_snps
Retrieving data from multiple tables using a JOIN
This query will retrieve the snp_id and ancestral_alleles columns from the dbSNP table _loc_snp_summary for those SNPs derived from entries in the Enter Genomic Features section. A MySQL INNER JOIN statement is used to describe how the two tables are to be combined. The ON keyword is used to indicate a matching condition. In this case, the column snp_id is common to both tables and can be used to match rows and combine columns. When multiple tables are involved it may be necessary to specify both the table and column using table.column notation as shown in the query below. Note that snp_id occurs in both tables and MySQL would return an error message if the table were not specified to remove the ambiguity.
SELECT _loc_snp_summary.snp_id,
_loc_snp_summary.ancestral_alleles
FROM _loc_snp_summary
INNER JOIN my_snps ON (_loc_snp_summary.snp_id = my_snps.snp_id)
Using aliases to simplify queries
Writing out the table names in queries can be laborious. Therefore MySQL allows you to specify a short alias for the name of a table. In the following query the AS keyword is used to specify table aliases. Actually the AS keyword may be omitted, but may improve readability.
SELECT LSS.snp_id,
LSS.ancestral_alleles
FROM _loc_snp_summary AS LSS
INNER JOIN my_snps AS MS ON (LSS.snp_id = MS.snp_id)
Joining several tables
More than two tables may joined together as illustrated by the example below. Note that the column snp_id, which is sometimes a unique identifier for a table, is not unique in the dbSNP table b134_SNPContigLocusId_37_2. This is because the variant may map to multiple contigs and gene transcripts and functional data may exist for separate alleles. It may therefore be necessary to include columns such as ctg_id, contig_acc and allele in order to explain the multiplicity.
SELECT LSS.snp_id,
LSS.ancestral_alleles,
SNP.avg_heterozygosity,
SCLI.locus_symbol,
SCLI.ctg_id,
SCLI.contig_acc,
SCLI.allele
FROM _loc_snp_summary AS LSS
INNER JOIN my_snps AS MS ON (LSS.snp_id = MS.snp_id)
INNER JOIN SNP ON (LSS.snp_id = SNP.snp_id)
INNER JOIN b134_SNPContigLocusId_37_2 AS SCLI ON (LSS.snp_id = SCLI.snp_id)
Counting rows
The MySQL COUNT function can be used to compute the number of rows in a table. The following query counts the number of SNPs in the table SNP - a dbSNP table with summary information on each unique reference SNP or rs ID. The query results in a table with a single column named COUNT(*) and a single row showing the number of SNPs.
SELECT COUNT(*) FROM SNP
Aggregate queries - counting the number of SNPs per gene
One of the powerful features of direct MySQL querying is the ability to perform aggregate queries. The following query counts the number of SNPs associated with each gene symbol in the column rep_gene_symbol. Here, the MySQL keywords GROUP BY are used to group rows together by the values of a specified column, in this case the gene symbols for the SNPs from Enter SNPs. The COUNT function then applies to the groups. Other MySQL functions that may be applied to groups include MIN, MAX, AVG and SUM.
SELECT LSS.rep_gene_symbol AS "Gene Symbol",
COUNT(*) AS "Number of SNPs"
FROM my_snps MS
INNER JOIN _loc_snp_summary AS LSS ON (MS.snp_id = LSS.snp_id)
GROUP BY LSS.rep_gene_symbol
ORDER BY COUNT(*) DESC
Note that in this example we have used the AS keyword after column specifications in the SELECT statement. This has the effect of naming the columns in the results of the query. This is useful when functions and other complex expressions are used in the query.
Aggregate queries - creating a list of submitters
Another useful MySQL aggregate function is GROUP_CONCAT which will combine the values of a column specified in a GROUP BY clause into a comma-separated list. When used with the DISTINCT keyword, it will remove duplicate values in the list. The following query will create for each SNP a list of submitters derived from the column _loc_submissions.handle.
SELECT MS.snp_id,
GROUP_CONCAT(DISTINCT LS.handle) AS Submitters
FROM my_snps AS MS
INNER JOIN _loc_submissions AS LS ON (MS.snp_id = LS.snp_id)
GROUP BY MS.snp_id