Documentation Tools
This describes the BioQ database documentation tools, sometimes referred to as dbDoc, including the tracing diagrams and the use of the Biologic-Experiment-Result (BERT) relational model. The code can be found on our . For example, see the documentation on our implementation the HapMap relational database for Public Release 28.
Contents
Files
back_trace.php
Create the back trace diagram, based on the Biologic-Experiment-Results (BERT) model, using dbdoc_util.pl.
column.php
Details on a specific column.
dbdoc_util_trace.opt
These options should be specified in the main options file.
db.php
Details on a specific database.
dbdoc_util_trace.opt
Options for running dbdoc_util.pl.
flow_group.php
Details on a specific flow group from the BERT model.
jqgrid_query_column*.php
This supplied the data to jqGrid options created in JavaScript. For example, to show a list of tables in db.php, in js/db.js we have the code
// List of tables
$('#dbDocTableTable-jqgrid').jqGrid({
url:'jqgrid_query_table.php',
postData: {db_name: dbDocDbName, filter: dbDocFilter},
datatype: 'xml',
mtype: 'GET',
pager: '#dbDocTableTable-jqgridPager',
altRows: true,
altclass: 'dbDocjqGridZebra',
height: 'auto',
//rowNum: 1000000,
rowNum: 30,
rowList: [10,20,30,50,100],
//caption: 'Tables',
hidegrid: false,
sortname: 'table_name',
sortorder: 'asc',
viewrecords: true,
pagerpos: 'left',
recordpos: 'center',
colNames: ['Name','Description','Rows','Size (Mb)'],
colModel: [
{name: 'Name', index: 'table_name', width: '225', align: 'left'},
{name: 'Description', index: 'short_descrip', width: '500', align: 'left'},
{name: 'Rows', index: 'rows', width: '90', align: 'right', formatter: 'integer', formatoptions: {thousandsSeparator: ',', defaultValue: '-' }},
{name: 'Table Size (Mb)', index: 'table_size', width: '80', align: 'right', formatter: 'integer', formatoptions: {thousandsSeparator: ',', decimalPlaces: 2, defaultValue: '-' }}
]
});
This uses jqgrid_query_table.php to get the data from MySQL. The data is sent via Ajax in XML format.
link_functions.php
Need to move this code to bioq/common.php.
process.php
Details on a specific process from the BERT model.
process_trace.php
This may be obsolete, it was originally used to show a large process trace diagram after clicking a thumbnail.
search.php
This page shows the results of a search.
search_terms.php
This is used for the jQuery UI autocomplete widget. It sends the data for the autocomplete entries via Ajax.
search_terms_ui.sql
This script is obsolete and needs to be replaced by something more sophisticated that populates the table `search_terms` from existing dbDoc databases. See http://deku.psych.wucon.wustl.edu:8081/browse/BIOQ-18.
session.php
This sets up PHP sessions for those pages that need it.
table.php
Details on a specific table.
table_trace.php
This may be obsolete, it was originally used to show a large table trace diagram after clicking a thumbnail.
tag.php
Details on a specific tag.
trace_table_legend.gif
The legend shown in the back trace section of BioQ::Documentation.
This needs to be updated whenever the code for displaying diagrams is updated. See http://deku.psych.wucon.wustl.edu:8081/browse/BIOQ-9.
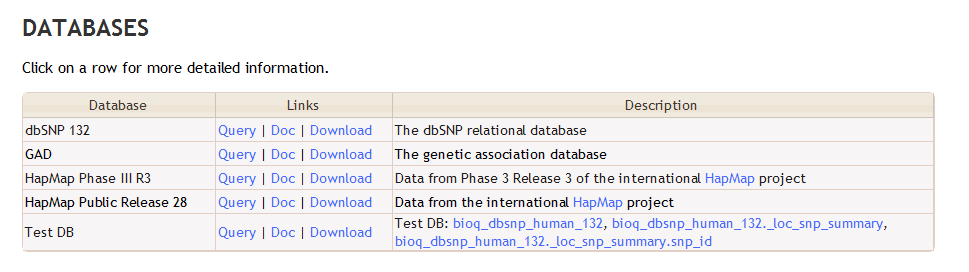
Including a test database in the display
In order to show a test database in the main table of databases, in bioq/dbdoc/jqgrid_query_db.php, search for 'bioq_test_db' and follow the commented instructions to show some test documentation. This is useful for testing out macros and things, such as _tbl_link().
When done correctly, you should see something like this:
which shows this entry from the dbdoc `db` database:
Test DB: _db_link(bioq_dbsnp_human_132), _db_tbl_link(bioq_dbsnp_human_132._loc_snp_summary), _db_tbl_col_link(bioq_dbsnp_human_132._loc_snp_summary.snp_id)