Description
The Perl script dbdoc_utilities.pl is used to populate the MySQL databases for database documentation in BioQ::Documentation. It also draws the "tracing" diagrams using the BERT model.
Documentation Status
- Think about separate sections for dbdoc_util.pl and database stuff
- Need to document dbdoc_util.pl
Tasks
Documentation Tasks
Contents
Directories and Links
Item |
Location |
Notes |
|---|---|---|
Subversion |
svn://deku.psych.wucon.wustl.edu/bioq/trunk/web/perl/dbdoc |
|
Scott's working directory |
/projects/bioinf/ssaccone/dbdoc_util |
Uses the older Subversion project dbdoc_util. |
dbdoc_util.pl
This program creates, configures and maintains the dbDoc MySQL database (currently bioq_dbdoc_1). It will initialize the documentation database, copy documentation from one database to another, update documentation from an XML file, dump the database, and will also generate process flow diagrams using GraphViz.
db_doc_model.mwb
A MySQL Workbench file containing the schema and ER diagrams. The diagrams below were taken directly from this file.
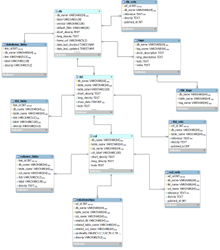
The core model
This entity-relationship (ER) diagram shows the core relational model for our database documentation (from subversion revision 70).
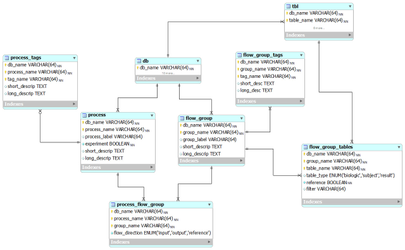
The BERT model
Our Biologic-Experiment-Results (BERT) relational model (from subversion revision 70).
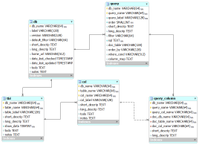
The queries model
Our model for storing queries in the documentation database. This information is used to populate the BioQ queries for each database.
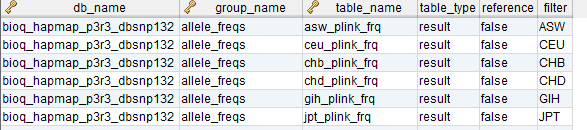
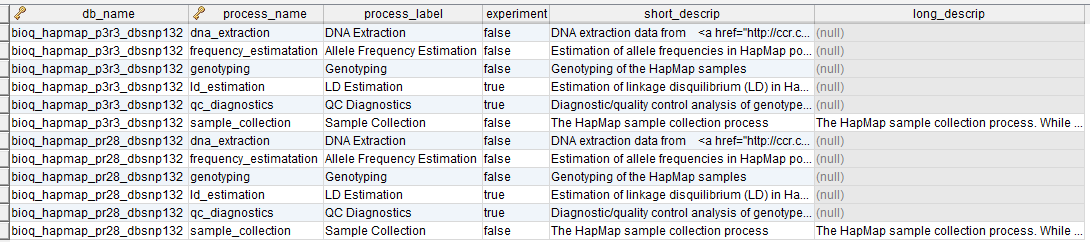
The BioQ documentation database
The BioQ::Documentation (dbDoc) features, which allow investigators to browse genomic database documentation, are based on a single MySQL database (TODO: specify PHP variable name). The current documentation database is bioq_dbdoc_1. Some ER diagrams for specific models in the database are shown above.
dbDoc Tables
db, tbl and col
These provide descriptions of database, tables and columns, respectively, in the BioQ genomic databases.
db_refs, tbl_refs and col_refs
Bibliographic references.
database_links, column_links, tbl_links
Web links.
To do: we should rename this to db_links, etc. They are probably not reference in code very much, if at all. See http://deku.psych.wucon.wustl.edu:8081/browse/BIOQ-21.
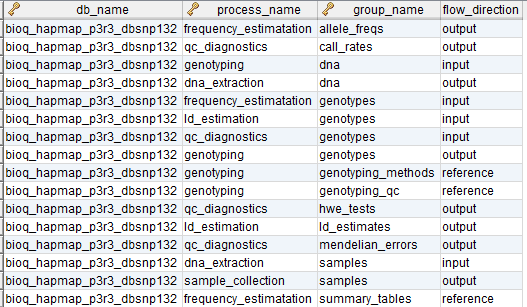
flow_group
In the BERT model, a flow group is a group of tables that can be input or output for a specific process.
flow_group_tables
The tables in a flow group.
flow_group_tags
Tags that describe a flow group. These are not the same as filters.
process
A process in the BERT model.
process_flow_group
A group of tables, columns and/or databases which are input/output for a process or experiment in the BERT model. It's possible to have multiple groups with the same name for different processes. Each process must have a group and groups may be singletons.
process_tags
Tags that describe a process. These can also be used as filters. No entries as of version 70.
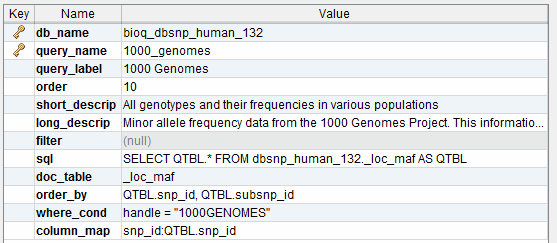
query
A query used in the BioQ web application.
query_column
This is used in Process.pm to look up information about columns being queried.
It's actually not clear what the purpose of query_column is. Need to make sure this is actually being used, and for what.
reference
Reference table showing things like Subversion revision number.
relationships
For recording and describing relationships between tables in genomic databases. No entries as of version 70.
results_tables
For each table with table_type='result', determine if it can be traced to subjects and/or biologics.
search_terms
Items used in the the dbDoc autocomplete search tool.
Need to improve how this table is populated. See http://deku.psych.wucon.wustl.edu:8081/browse/BIOQ-18.
tags
Tags are used to group tables into categories. The 'tags' table contains names and descriptions of the tags.
tbl_tags
This records which tags go with which tables.
Updating the dbDoc schema
- Dump the existing database
- Use mysql --complete-insert --no-create-db --no-create-info --verbose
- See /projects/bioinf/ssaccone/dbdoc_util/dump_dbdoc_data.sh
- If necessary, create the schema file (current using MySQL Workbench with the file db_doc_model.mwb)
- The schema file is currently named db_doc_model.sql
- Note that the schema file should not have a 'USE' statement - instead use the dbdoc_util.pl dbdoc-db option.
- Run dbdoc_util.pl initdb
- You may also use the predefined script template_init.sh and the correpsonding options file template_init.opt
- Load the original dumped database
- See also the script /projects/bioinf/ssaccone/dbdoc_util/update_schema.sh
Initializing documentation for a single database
This is used to take an existing genomic database, such as HapMap, and set up entries in the dbDoc database for all the tables and columns in the database. This will ensure that entries for the different tables and columns exist so that more detailed documentation can be added.
Example: see /projects/bioinf/ssaccone/hapmap/dbdoc/hapmap_p3r3_dbsnp132.sh
- Run dbdoc_util.pl initdoc (with options) to set up dbDoc for a specified existing genomic MySQL database
- Run dbdoc_util.pl updatedoc --dbdoc-xml-file=<file> to read XML documentation
- Suggestion: break up XML documentation into multiple files, such as a "core" file that does not change often, and a another file that might describe what the recent changes are. This is currently done for the HapMap databases.
Modifying documentation
- Deletions: at the moment (subversion 29) a row can be deleted from the db table and this will propagate throughout the database.
- Updates: at the moment (subversion 29) changes do not cascade well due to a circular foreign key in the process/flow_group tables. To change the name of a database the docoumentation should be re-loaded from XML.