Some queries refer to certain genomic features such as SNPs and genes. If you specify a list of SNPs, genes or and/or genomic regions in the Enter Genomic Features section, BioQ will retrieve results only for those features. For example, if a SNP-related query is run and a genomic region is specified, only SNPs in that region will be retrieved. Different kinds of features, such as SNPs and genes, can be combined in the same query.
Features must be separated by carriage returns. They may be typed directly into the the Enter Genomic Features section or uploaded as a file. An optional comment may be provided by inserting a comma after the feature followed by the comment. The status indicator in the upper right of the features window turns green after a query and indicates that your features are now saved on the server. If you do not change these features, it will remain green and those features can be used for future queries. This saves the time of processing the features. The indicator turns red when you modify the queries. There is currently no undo mechanism, although your web browser may provide such a feature.
...
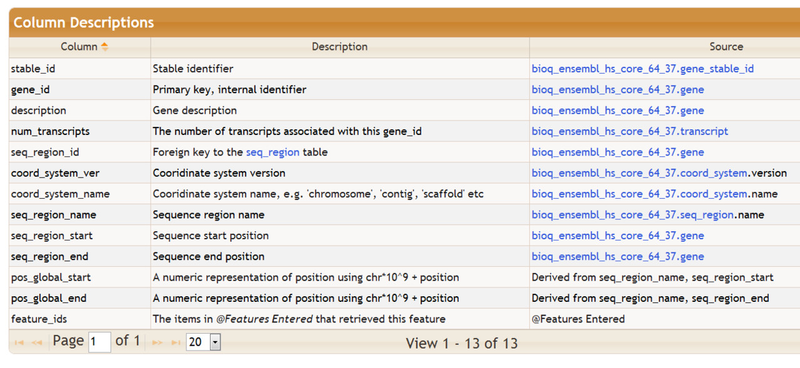
Brief descriptions of the columns in the feature table are also provided, as shown below. The Source column describes, when possible, the data source for that particular column in the feature table. The data source is usually a table in one of the BioQ databases , and is often linked to more detailed information about the source.
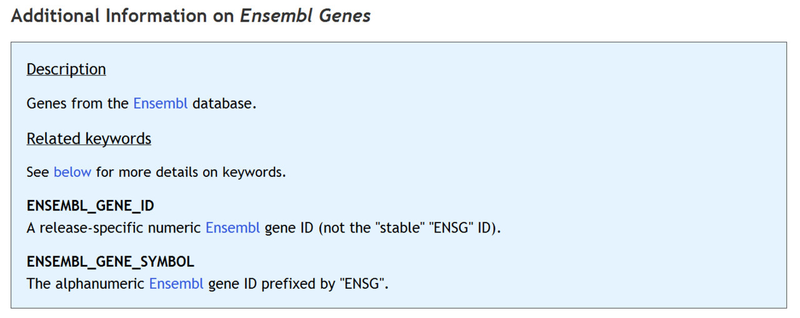
Additional information on the selected feature table, including the list of keywords that can be used to populate the table, are can be found below below the column descriptions:
Interpopulation of Features
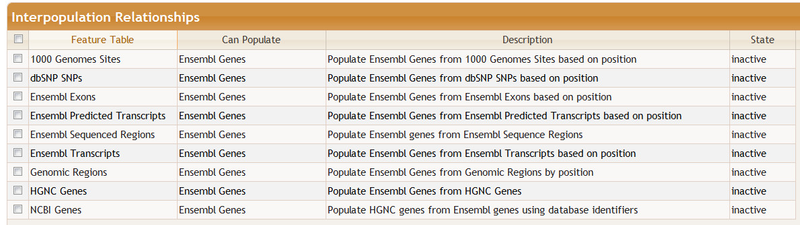
Below the feature tables is the Interpopulation Relationships section. This section describes how the selection of certain features, such as genomic regions, can automatically trigger the selection of other features, such as genes in the region. In other words, the Genomic Regions feature will populate Ensembl Genes and we call this an interpopulation relationship.
...
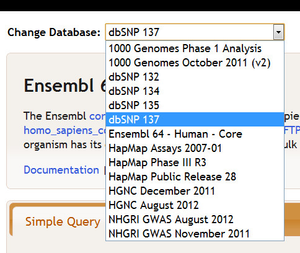
For example, if we switch from Ensembl 64 - Human - Core to dbSNP 137:...
Then ...then some interpopulations will change. In particularly; in particularly, those that populate Ensembl Genes become inactive.:
You may activate or deactive a relationship. Clicking on a row some provides configuration options plus some additional documentation. You may select multiple rows for activation/deactivation by checking the boxes on the left.
...
You may change the order in which relationships are executed. For example, to find exonic 1000 Genomes variants in a region using Ensembl data you may want to ensure that Ensembl Exons populates 1000 Genomes Sites after Genomic Regions populates Ensembl Exons.
...
The bottom of the Genomic Features sectrion contains detailed information on how to specify features in the Enter Genomic Features section. Features are entered mainly by using keywords. A table of these keywords is given where - detailed information may be found by clickingn clicking on row. For some features the keywords are optionoptional: for example, see the keywords DBSNP_SNP_ID and NCBI_GENE_SYMBOL.
...