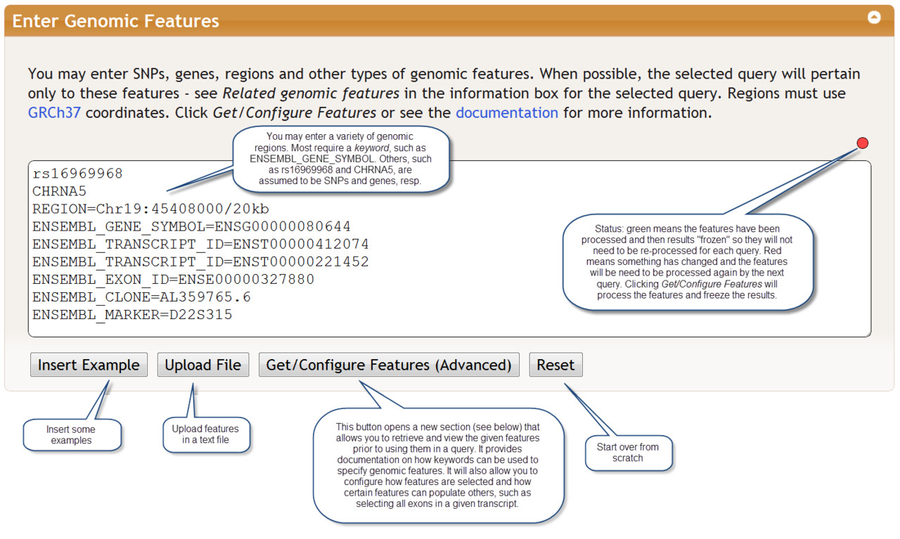
Some queries refer to certain genomic features such as SNPs and genes. If you specify a list of SNPs, genes or and/or genomic regions in the Enter Genomic Features section, BioQ will retrieve results only for those features. For example, if a SNP-related query is run and a genomic region is specified, only SNPs in that region will be retrieved. Different kinds of features, such as SNPs and genes, can be combined in the same query.
Features must be separated by carriage returns. They may be typed directly into the the Enter Genomic Features section or uploaded as a file. An optional comment may be provided by inserting a comma after the feature followed by the comment. The status indicator indicator in the upper right of the features window turns green after a query and indicates that your features are now saved on the server. If you do not change these features, it will remain green and those features can be used for future queries. This saves the time of processing the features. The indicator turns red when you modify the queries. There is currently no undo mechanism, although your web browser may provide such a feature.
Subtopics
| Table of Contents | ||
|---|---|---|
|
Entering Variants
dbSNP IDs
Specific genetic variants are entered dbSNP IDs. The "rs" (reference SNP) prefix is optional.
| Code Block | ||||
|---|---|---|---|---|
| ||||
[SNP=][rs]<numeric dbSNP ID> |
| Code Block | ||||
|---|---|---|---|---|
| ||||
16969968
rs16969968
SNP=16969968
SNP=rs16969968 |
Entering Genes
When a gene is specified, any SNP-related query will be applied to all SNPs found in this gene using the SNP/gene transcript relationships from the latest build of dbSNP (currently 134). Genes are specified using alphanumeric NCBI Entrez Gene symbols, like CHRNA5, or numeric Entrez Gene IDs, like 1138.
NCBI Entrez Gene Symbols (alphanumeric)
keyword: GENE
aliases: NCBI_GENE_SYMBOL, ENTREZ_GENE_SYMBOL, ENTREZ_GENE
A single gene may be specified using the NCBI Entrez Gene symbol. The GENE= prefix may optionally be used for clarity.
| Code Block | ||||
|---|---|---|---|---|
| ||||
[GENE=]<alphanumeric NCBI Entrez Gene symbol> |
| Code Block | ||||
|---|---|---|---|---|
| ||||
CHRNA5
GENE=CHRNA5 |
NCBI Entrez Gene IDs (numeric)
keyword: GENE_ID
aliases: NCBI_GENE_ID, ENTREZ_GENE_ID
Gene IDs must use the GENE_ID= prefix in order to distinguish them from dbSNP SNP IDs, which are the default numeric ID.
| Code Block | ||
|---|---|---|
| ||
GENE_ID=<numeric NCBI Entrez Gene ID> |
| Code Block | ||||
|---|---|---|---|---|
| ||||
GENE_ID=1138 |
Using search terms to specify genes
An entire set of genes may be specified using a the ENTREZ_GENE_QUERY= prefix. Multiple search terms may be grouped with the AND operator and phrases may be placed in double quotes.
| Code Block | ||||
|---|---|---|---|---|
| ||||
ENTREZ_GENE_QUERY="cholinergic receptor"
ENTREZ_GENE_QUERY=nicotinic AND beta AND muscle |
Entering Genomic Regions
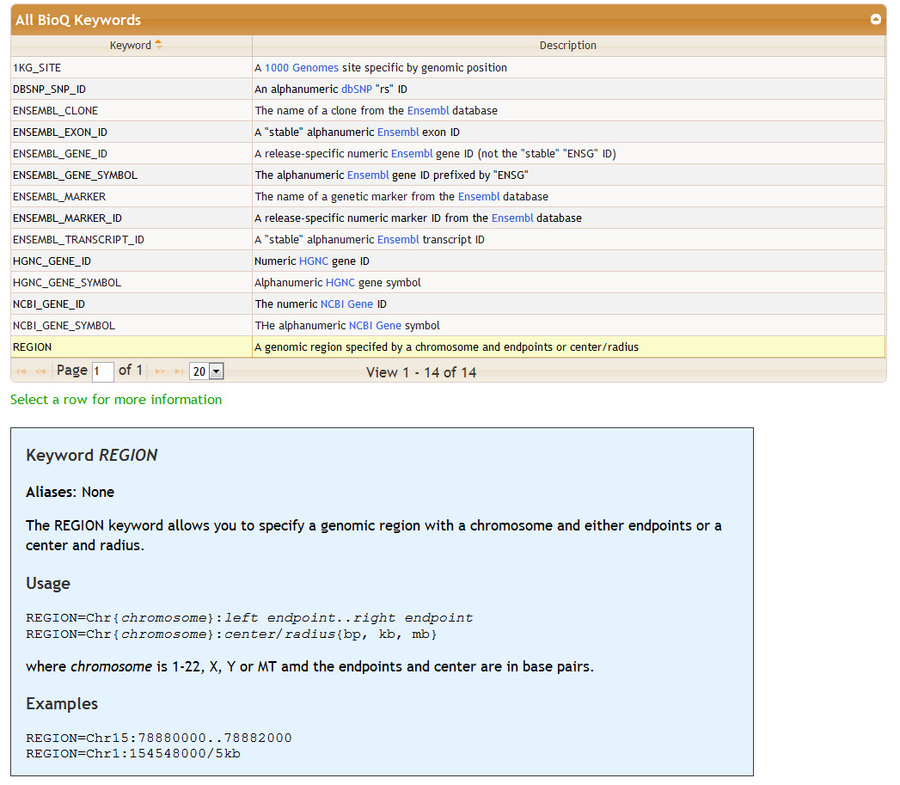
keyword: REGION
Genomic regions are specified with a chromosome and starting and ending positions in base pairs.
| Code Block | ||||
|---|---|---|---|---|
| ||||
REGION=Chr<chromosome:1-22,X,Y>:<start position (bp)>..<end position (bp) |
| Code Block | ||||
|---|---|---|---|---|
| ||||
REGION=Chr15:78880000..78882000 |
Ensembl Features
| Info |
|---|
The Ensembl database is still in progress. |
Ensembl Clones
keyword: ENSEMBL_CLONE
A clone from the Ensembl Human Core database.
| Code Block | ||
|---|---|---|
| ||
ENSEMBL_CLONE=<alphanumeric clone ID> |
| Code Block | ||
|---|---|---|
| ||
ENSEMBL_CLONE=AL359765.6 |
The clone ID is matched against the column seq_region.name in the Ensembl Human Core database.
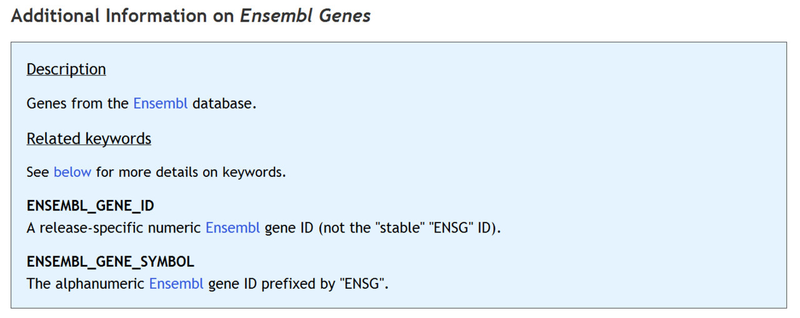
Ensembl Genes
keyword: ENSEMBL_GENE_SYMBOL
aliases: ENSEMBL_STABLE_GENE_ID
| Code Block | ||
|---|---|---|
| ||
ENSEMBL_GENE_SYMBOL=<alphanumeric stable Ensembl gene ID> |
| Code Block | ||
|---|---|---|
| ||
ENSEMBL_GENE_SYMBOL=ENSG00000080644 |
Ensembl Transcripts
keyword: ENSEMBL_TRANSCRIPT_ID
aliases: ENSEMBL_STABLE_TRANSCRIPT_ID
| Code Block | ||
|---|---|---|
| ||
ENSEMBL_TRANSCRIPT_ID=<alphanumeric stable Ensembl gene ID> |
| Code Block | ||
|---|---|---|
| ||
ENSEMBL_TRANSCRIPT_ID=ENST00000412074 |
...
Checking How BioQ Interpreted Your Features
After executing a query, at the bottom of the query results page you will find an Additional Information section. It provides some details on how BioQ processed the items you entered in the Genomic Features section. Suppose we entered the following features (note that we included optional comments followed by comma after each feature):
The Summary tab provides some summary information about your features and will tell you if any errors were found.
The SNP/Gene/Region Queries section displays the actual features, the optional comments, and how many variants and genes were retrieved for each features. It also shows the exact SQL WHERE statement that was used.
The SNPs section displays the actual SNPs discovered from genomic features entered.
The Genes section displays the genes resulting from the features entered.
...
Working with Features
When you click the Get/Configure Features button any features you have entered will be processed and the following section appears (you do not have to enter features to view this section):
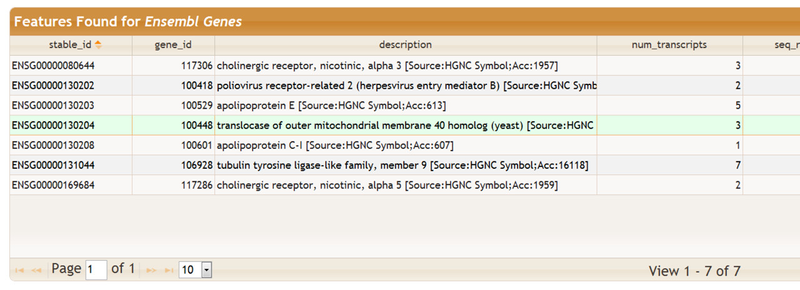
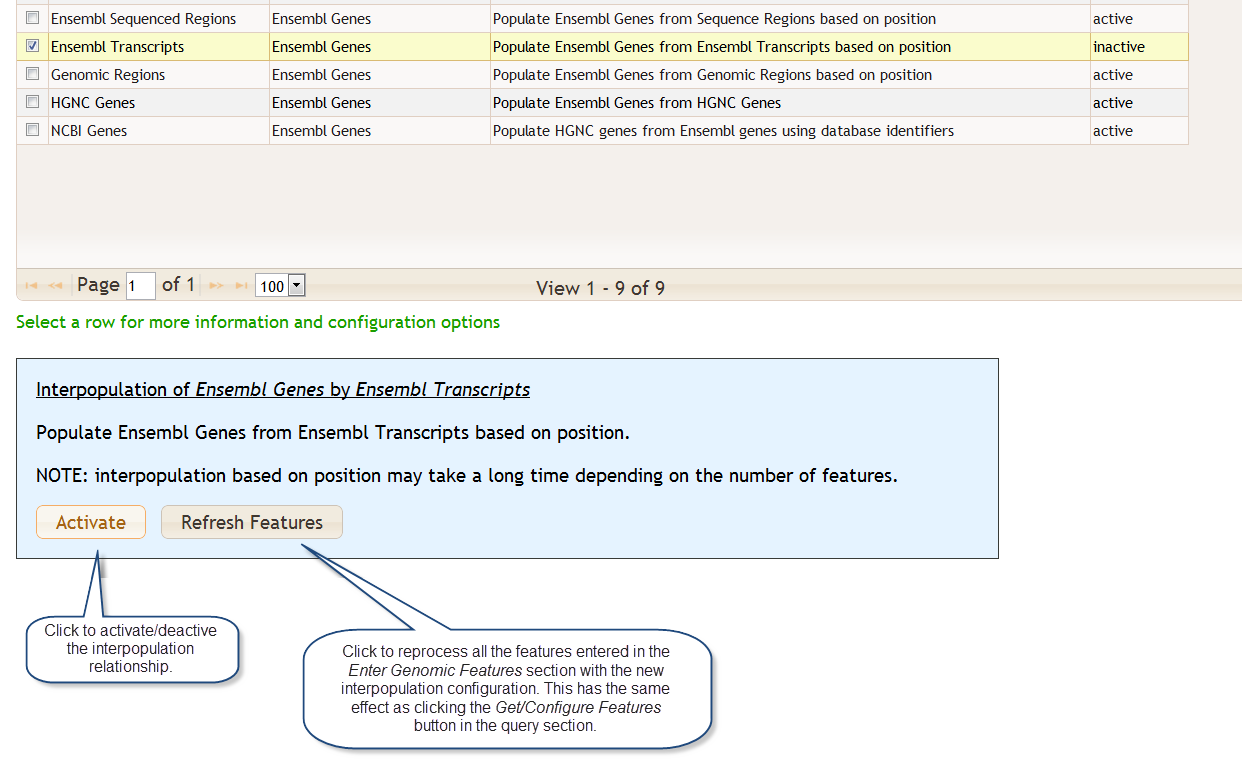
The table Features Found shows the genomic features that were retreived based on the items that were entered in the Enter Genomic Features section. The results are groups into tables called feature tables. You may click on a feature table to view the data it contains plus some documentation. Here we clicked on the Ensembl Genes feature table:
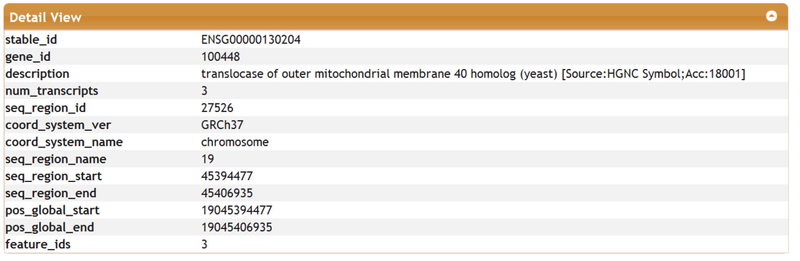
This a partial view of this table as it contains several columns. You may select an individual row for a detail view:
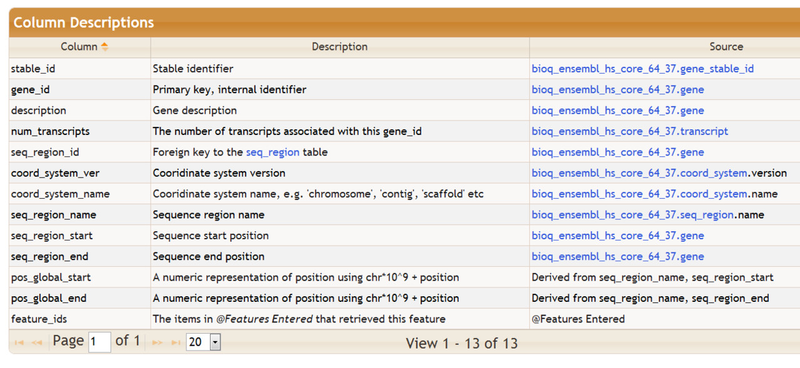
Brief descriptions of the columns in the feature table are also provided, as shown below. The Source column describes, when possible, the data source for that particular column in the feature table. The data source is usually a table in one of the BioQ databases and is often linked to more detailed information.
Additional information on the selected feature table, including the list of keywords that can be used to populate the table, can be found below the column descriptions:
Interpopulation of Features
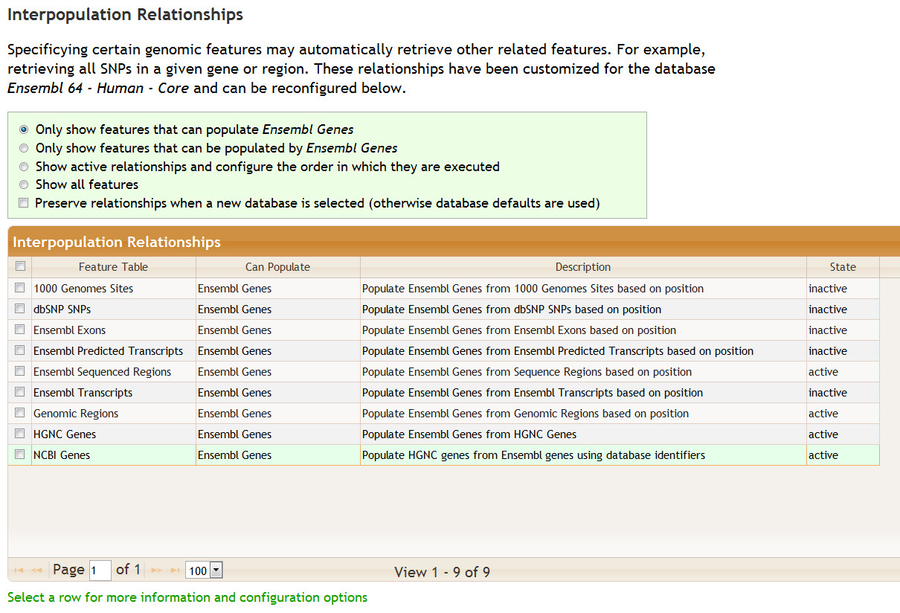
Below the feature tables is the Interpopulation Relationships section. This section describes how the selection of certain features, such as genomic regions, can automatically trigger the selection of other features, such as genes in the region. In other words, the Genomic Regions feature will populate Ensembl Genes and we call this an interpopulation relationship.
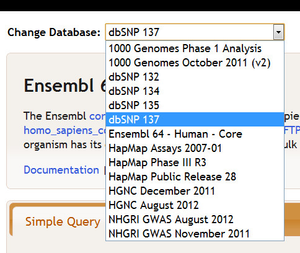
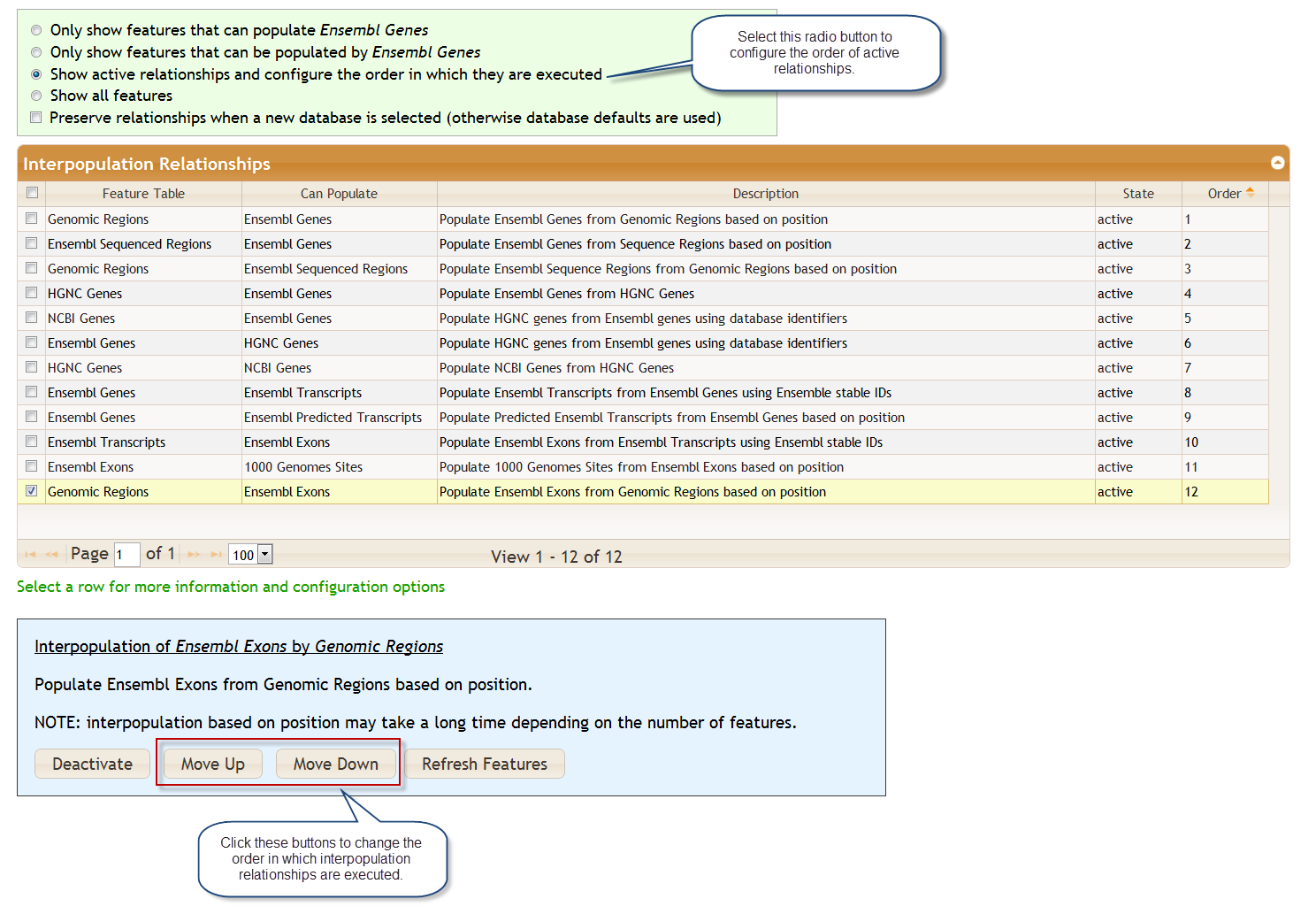
There are many possible interpopulation relationships. Clicking on the various radio buttons controls how many are shown. The relationships may be active or inactive. The default configuration depends on the database selected. For example, if you are working with the dbSNP database you may not want to have genomic regions populate Ensembl genes, particularly because this will require additional processing time. If you select a new database at the top of the query page, the interpopulation relationships will change.
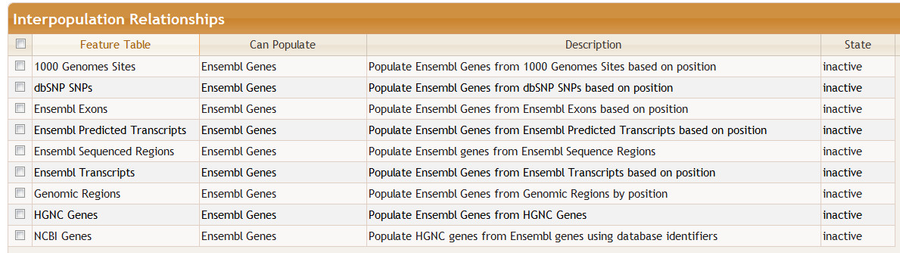
For example, if we switch from Ensembl 64 - Human - Core to dbSNP 137...
...then some interpopulations will change; in particularly, those that populate Ensembl Genes become inactive:
You may activate or deactive a relationship. Clicking on a row some provides configuration options plus some additional documentation. You may select multiple rows for activation/deactivation by checking the boxes on the left.
You may change the order in which relationships are executed. For example, to find exonic 1000 Genomes variants in a region using Ensembl data you may want to ensure that Ensembl Exons populates 1000 Genomes Sites after Genomic Regions populates Ensembl Exons.
Using Keywords to Specify Features
The bottom of the Genomic Features sectrion contains detailed information on how to specify features in the Enter Genomic Features section. Features are entered mainly by using keywords. A table of keywords is given - detailed information may be found by clicking on row. For some features the keywords are optional: for example, see the keywords DBSNP_SNP_ID and NCBI_GENE_SYMBOL.
Using Feature Tables in Queries
Some queries, such as the Genes query in the database Ensembl 64 - Human - Core, can be limited by certain feature tables:
If you do not enter any features, the query will retrieve a random set of data taken in whatever order the data exists in the database. If you enter features related to Ensembl Genes, such as by explicitly using the ENSEMBL_GENE_SYMBOL keyword or by using an interpopulation relationship such as getting the genes from regions, then the results of the Genes query will be limited to those features.
| Info |
|---|
The following is technical information on how feature tables are used in MySQL queries |
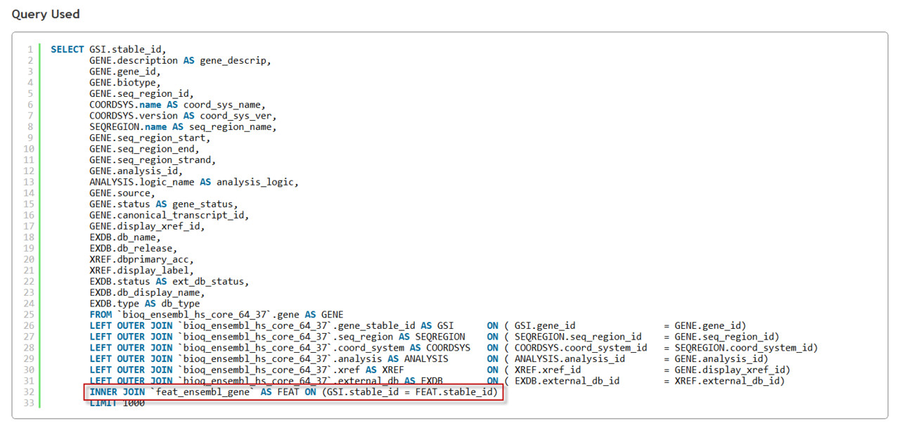
After the query is executed, if the query did use the Ensembl Genes feature table you will see a reference to its MySQL table feat_ensembl_gene in the Query Used section:
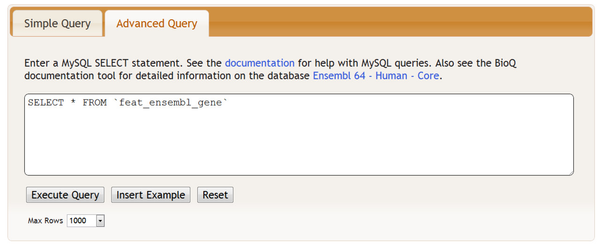
You may also access the feature tables in the Advanced Query section:
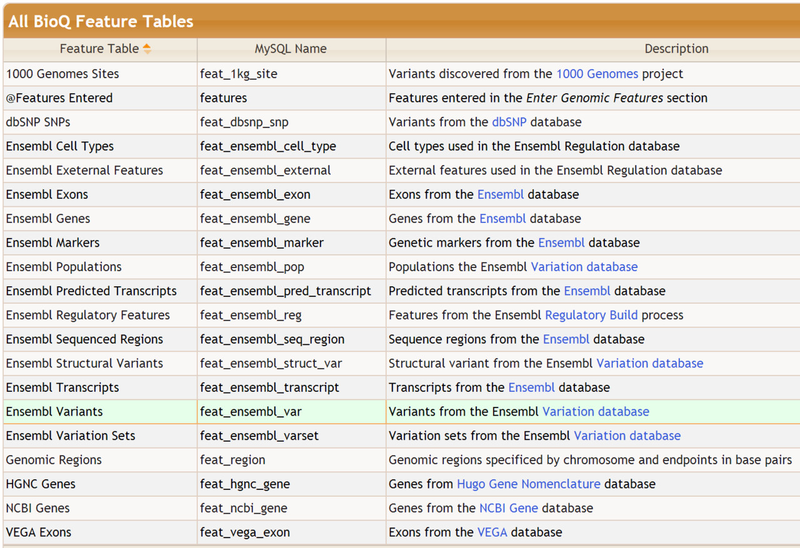
Complete List of Features
The complete list of features at the time of writing:
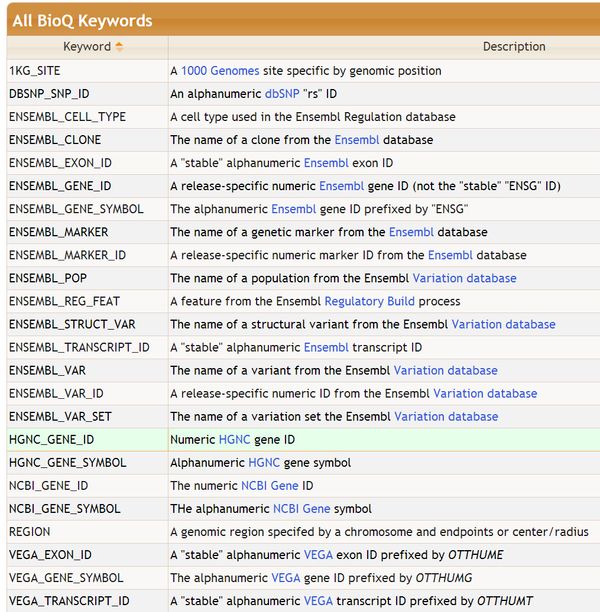
Complete List of Keywords
The complete list of features at the time of writing: