...
The
...
BioQ
...
web
...
application
...
uses
...
several
...
relational
...
database
...
models
...
to
...
provide
...
documentation
...
and
...
query
...
tools
...
for
...
the
...
various
...
genomic
...
databases.
...
All
...
schemas
...
and
...
ER
...
diagrams
...
can
...
be
...
found
...
in
...
the
...
...
...
document
...
on
...
our
...
Subversion
...
server.
...
Contents
| Table of Contents | ||
|---|---|---|
|
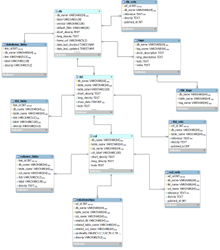
The core relational model
This entity-relationship (ER) diagram shows the core relational model for our database documentation (from subversion revision 70).
The biologic-experiment-result
...
(BERT)
...
relational
...
model
...
Our
...
BERT
...
relational
...
model
...
is
...
designed
...
to
...
document
...
a
...
genomic
...
database
...
by
...
tracing
...
the
...
experimental
...
source
...
of
...
the
...
data.
...
The
...
key
...
entities
...
in
...
the
...
models
...
are
...
the
...
biologics,
...
the
...
experiments
...
and
...
the
...
experimental
...
results
...
as
...
illustrated
...
in
...
the
...
following
...
diagram.
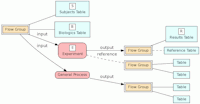
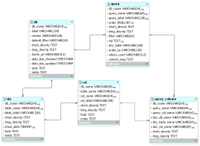
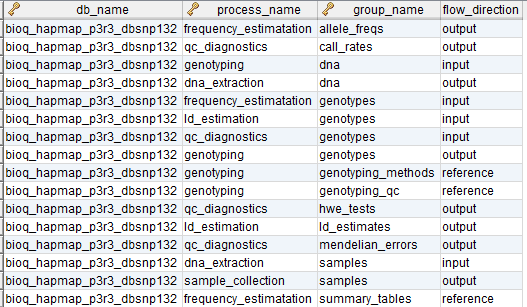
In the model, the tables in a relational database are grouped into flow groups that can input, output or reference material for the various experiments and processes that determined the data. Here is an example from our BioQ web application of the BERT model being applied to linkage disequilibrium (LD) data from the HapMap database. In BioQ, the diagram is interactive and can be used to navigate documentation and query the actual data.
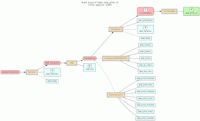
The following ER diagram shows the relational database implementation of the BERT model (from subversion revision 70, see the MySQL Workbench file ).
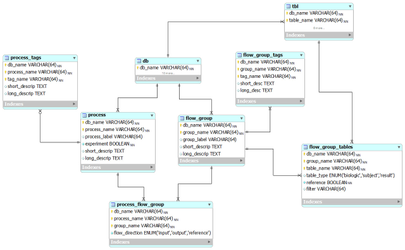
The queries model
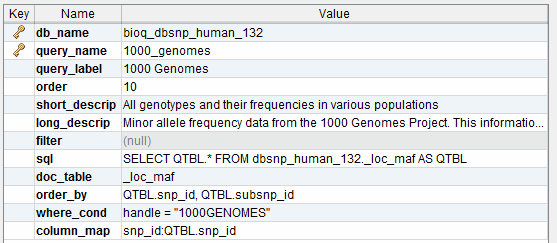
This is our model for storing queries in the documentation database. This information is used to populate the BioQ queries for each database.
Tables
db, tbl and col
These provide descriptions of database, tables and columns, respectively, in the BioQ genomic databases.
db_refs, tbl_refs and col_refs
Bibliographic references.
database_links, column_links, tbl_links
Web links.
flow_group
In the BERT model, a flow group is a group of tables that can be input or output for a specific process.
flow_group_tables
The tables in a flow group.
flow_group_tags
Tags that describe a flow group. These are not the same as filters.
process
A process in the BERT model.
process_flow_group
A group of tables, columns and/or databases which are input/output for a process or experiment in the BERT model. It's possible to have multiple groups with the same name for different processes. Each process must have a group and groups may be singletons.
process_tags
Tags that describe a process. These can also be used as filters. No entries as of version 70.
query
A query used in the BioQ web application.
query_column
This is used in Process.pm to look up information about columns being queried.
reference
Reference table showing things like Subversion revision number.
relationships
For recording and describing relationships between tables in genomic databases. No entries as of version 70.
results_tables
For each table with table_type='result',
...
determine
...
if
...
it
...
can
...
be
...
traced
...
to
...
subjects
...
and/or
...
biologics.
...
search_terms
...
Items
...
used
...
in
...
the
...
the
...
dbDoc
...
autocomplete
...
search
...
tool.
...
tags
Tags are used to group tables into categories. The 'tags'
...
table
...
contains
...
names
...
and
...
descriptions
...
of
...
the
...
tags.
...
tbl_tags
...
This
...
records
...
which
...
tags
...
go
...
with
...
which
...
tables.
...
Updating
...
the
...
documentation
...
schema
...
- Dump
...
- the
...
- existing
...
- database
...
- Use
...
- mysql
...
- --complete-insert
...
- --no-create-db
...
- --no-create-info
...
- --verbose
...
- See
...
- /projects/bioinf/ssaccone/dbdoc_util/dump_dbdoc_data.sh
...
- If
...
- necessary,
...
- create
...
- the
...
- schema
...
- file
...
- (current
...
- using
...
...
...
- with
...
- the
...
- file
...
- db_doc_model.mwb
...
- )
...
- The
...
- schema
...
- file
...
- is
...
- currently
...
- named
...
- db_doc_model.sql
...
- Note
...
- that
...
- the
...
- schema
...
- file
...
- should
...
- not
...
- have
...
- a
...
- 'USE'
...
- statement
...
- -
...
- instead
...
- use
...
- the
...
- dbdoc_util.pl
...
- dbdoc-db
...
- option.
...
- Run
...
- dbdoc_util.pl
...
- initdb
...
- You
...
- may
...
- also
...
- use
...
- the
...
- predefined
...
- script
...
- template_init.sh
...
- and
...
- the
...
- correpsonding
...
- options
...
- file
...
- template_init.opt
...
- Load
...
- the
...
- original
...
- dumped
...
- database
...
- See
...
- also
...
- the
...
- script
...
- /projects/bioinf/ssaccone/dbdoc_util/update_schema.sh
...
Initializing
...
documentation
...
for
...
a
...
single
...
database
...
This
...
is
...
used
...
to
...
take
...
an
...
existing
...
genomic
...
database,
...
such
...
as
...
HapMap,
...
and
...
set
...
up
...
entries
...
in
...
the
...
dbDoc
...
database
...
for
...
all
...
the
...
tables
...
and
...
columns
...
in
...
the
...
database.
...
This
...
will
...
ensure
...
that
...
entries
...
for
...
the
...
different
...
tables
...
and
...
columns
...
exist
...
so
...
that
...
more
...
detailed
...
documentation
...
can
...
be
...
added.
...
Example:
...
see
...
/projects/bioinf/ssaccone/hapmap/dbdoc/hapmap_p3r3_dbsnp132.sh
...
- Run
...
- dbdoc_util.pl
...
- initdoc
...
- (with
...
- options)
...
- to
...
- set
...
- up
...
- dbDoc
...
- for
...
- a
...
- specified
...
- existing
...
- genomic
...
- MySQL
...
- database
...
- Run
...
- dbdoc_util.pl
...
- updatedoc
...
- --dbdoc-xml-file=<file>
...
- to
...
- read
...
- XML
...
- documentation
...
- Suggestion:
...
- break
...
- up
...
- XML
...
- documentation
...
- into
...
- multiple
...
- files,
...
- such
...
- as
...
- a
...
- "core"
...
- file
...
- that
...
- does
...
- not
...
- change
...
- often,
...
- and
...
- a
...
- another
...
- file
...
- that
...
- might
...
- describe
...
- what
...
- the
...
- recent
...
- changes
...
- are.
...
- This
...
- is
...
- currently
...
- done
...
- for
...
- the
...
- HapMap
...
- databases.
...
Modifying
...
documentation
...
- Deletions
...
- :
...
- at
...
- the
...
- moment
...
- (subversion
...
- 29)
...
- a
...
- row
...
- can
...
- be
...
- deleted
...
- from
...
- the
...
- db
...
- table
...
- and
...
- this
...
- will
...
- propagate
...
- throughout
...
- the
...
- database.
...
- Updates
...
- :
...
- at
...
- the
...
- moment
...
- (subversion
...
- 29)
...
- changes
...
- do
...
- not
...
- cascade
...
- well
...
- due
...
- to
...
- a
...
- circular
...
- foreign
...
- key
...
- in
...
- the
...
- process/flow_group
...
- tables.
...
- To
...
- change
...
- the
...
- name
...
- of
...
- a
...
- database
...
- the
...
- docoumentation
...
- should
...
- be
...
- re-loaded
...
- from
...
- XML.