...
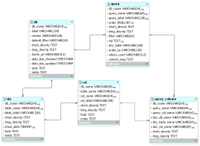
This entity-relationship (ER) diagram shows the core relational model for our database documentation (from subversion revision 70).
The biologic-experiment-result (BERT) relational model
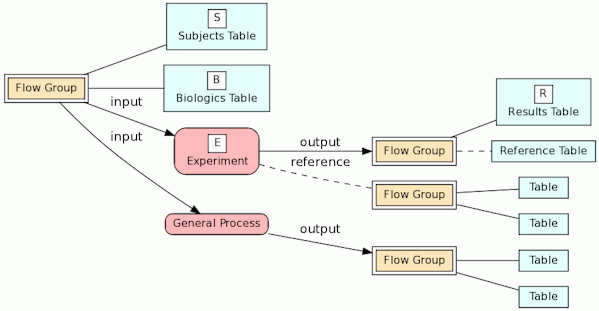
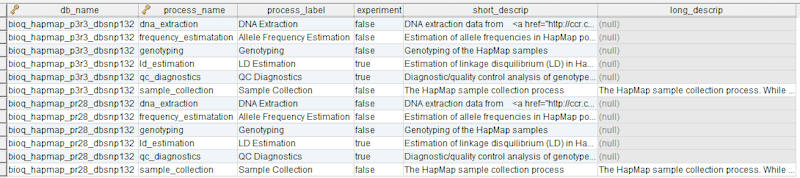
Our BERT relational model is designed to document a genomic database by tracing the experimental source of the data. The key entities in the models are the biologics, the experiments and the experimental results as illustrated in the following diagram.
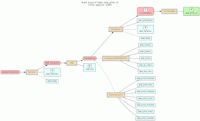
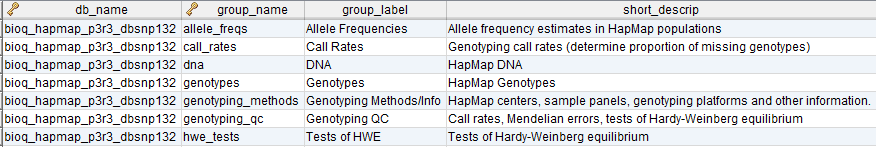
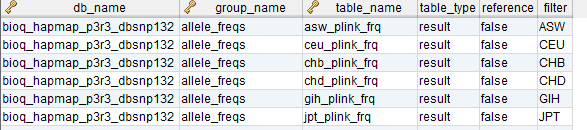
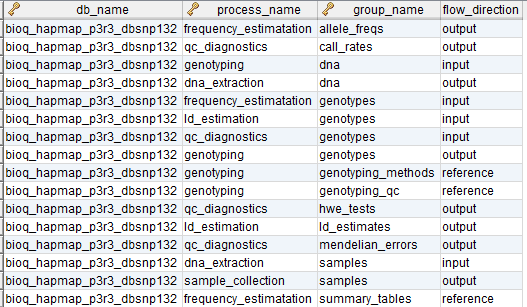
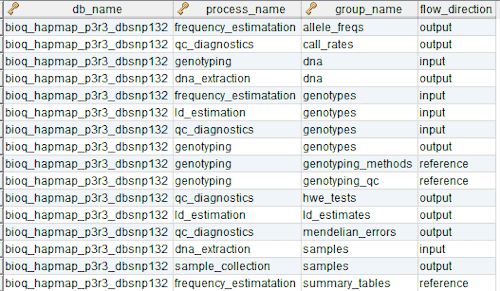
In the model, the tables in a relational database are grouped into flow groups that can input, output or reference material for the various experiments and processes that determined the data. Here is an example from our BioQ web application of the BERT model being applied to linkage disequilibrium (LD) data from the HapMap database. In BioQ, the diagram is interactive and can be used to navigate documentation and query the actual data.
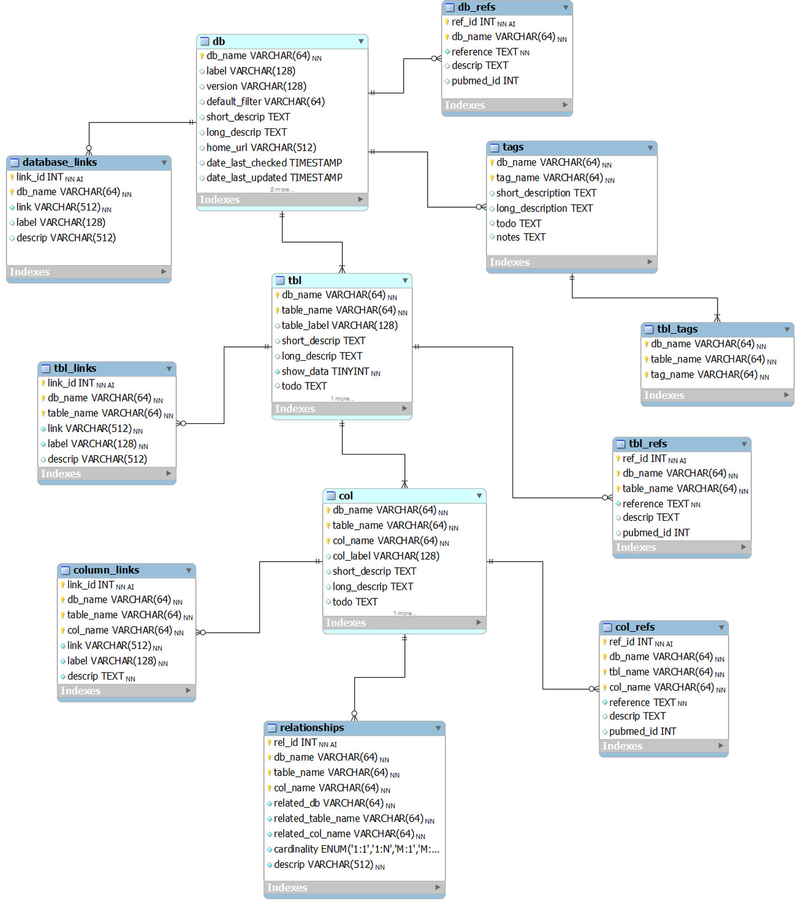
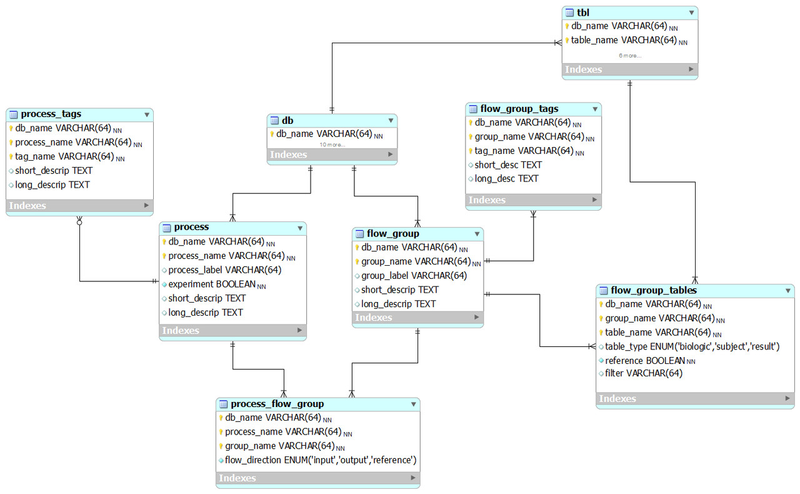
The following ER diagram shows the relational database implementation of the BERT model (from subversion revision 70, see the MySQL Workbench file ).
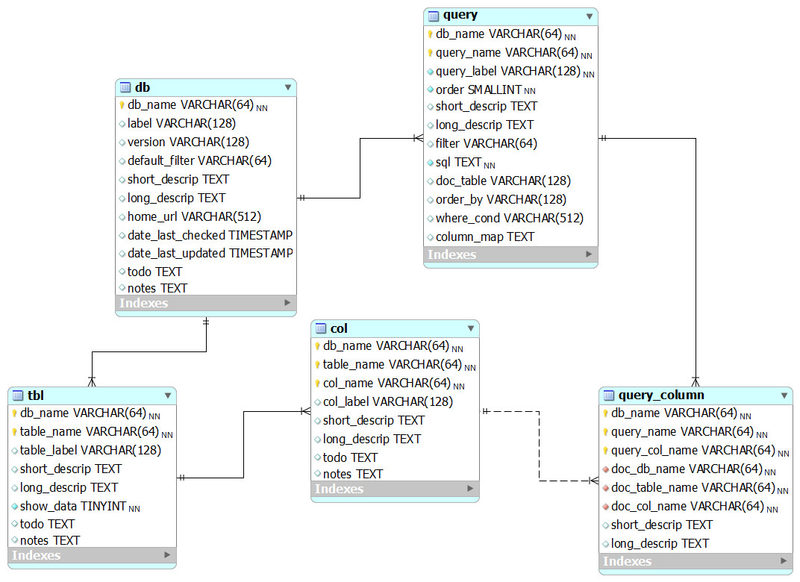
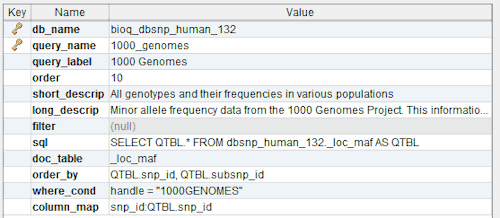
The queries model
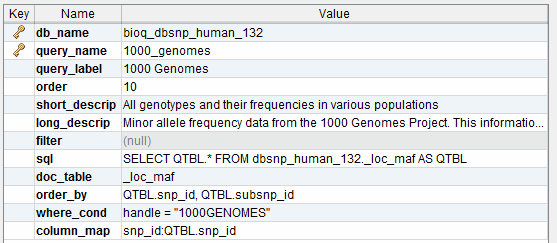
This is our model for storing queries in the documentation database. This information is used to populate the BioQ queries for each database.
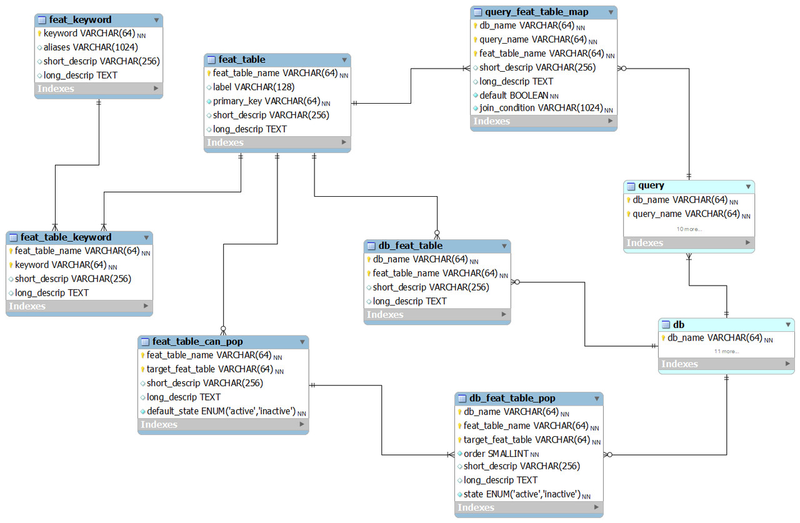
The features model
This is our model for querying genomic features such as genomic regions, gene transcripts, SNPs and other genetic variants. The way features are specified may depend on the database. For example, genes have different IDs in the NCBI, HUGO and Ensembl databases.
Features are specified in BioQ using keywords, such as SNP_ID and GENE_ID. There are usually defaults, such as a numeric feature being assumed to be a dbSNP rs ID and a alphanumeric string assumed to be a HUGO gene symbol.
...
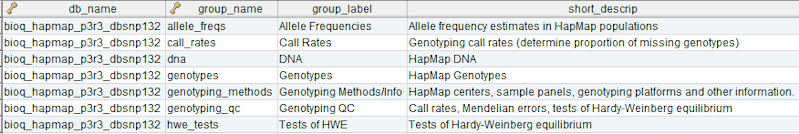
In the BERT model, a flow group is a group of tables that can be input or output for a specific process.
flow_group_tables
The tables in a flow group.
flow_group_tags
...
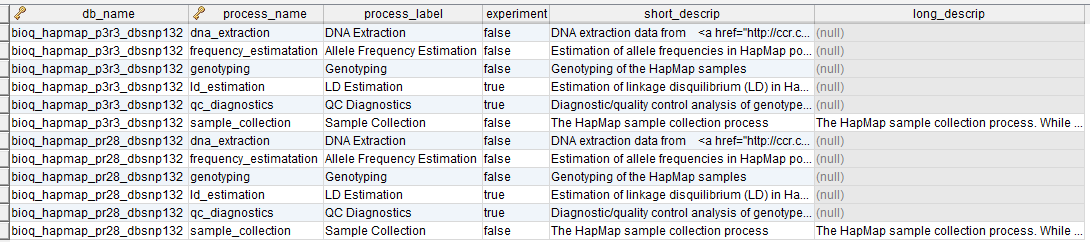
A process in the BERT model.
process_flow_group
A group of tables, columns and/or databases which are input/output for a process or experiment in the BERT model. It's possible to have multiple groups with the same name for different processes. Each process must have a group and groups may be singletons.
process_tags
Tags that describe a process. These can also be used as filters. No entries as of version 70.
...
A query used in the BioQ web application.
query_column
This is used in Process.pm to look up information about columns being queried.
...
This describes how specific feature tables are built using keywords. The keyword/feature table association is very important. It represents a process used to populate a feature table from a feature query (a feature specified on the BioQ query page). The descriptions in this table describe this process.
feat_table_can_pop
This explains how feature tables relate and interact with other feature tables. It contgains the other tables a feature table can populate. For example, the feature table feat_region can populate the table feat_dbsnp_snp via the relationship of the coordinate of a SNP being with the boundaries of a genomic region.
query_feat_table_map
This explains how to use feature tables once they are created. Specifically, it identifies the columns of a query that can be used to limit the query results by a feature table. For example, in the dbSNP database, the "SNP Summary" query can be limited to the features in the feat_dbsnp_snp feature table, as can any query that results in a dbSNP SNP ID column.
...
- Run dbdoc_util.pl initdoc (with options) to set up dbDoc for a specified existing genomic MySQL database
- Run dbdoc_util.pl updatedoc --dbdoc-xml-file=<file> to read XML documentation
- Suggestion: break up XML documentation into multiple files, such as a "core" file that does not change often, and a another file that might describe what the recent changes are. This is currently done for the HapMap databases.
Modifying documentation
- Deletions: at the moment (subversion 29) a row can be deleted from the db table and this will propagate throughout the database.
- Updates: at the moment (subversion 29) changes do not cascade well due to a circular foreign key in the process/flow_group tables. To change the name of a database the docoumentation should be re-loaded from XML.The documentation for individual databases in the runk/main/databases directory of the Subversion repository contain shell scripts for running dbdoc_util.pl. These scripts read the XML documentation files to the build the MySQL documentaiton database
- Documentation for an entire databases may be deleted by deleting row for that databases from the table db via MySQL cascade.
Locations of Documentation on Subversion
- Documentation for individual databases is in the trunk/main/databases directory of the Subversion repository.
- Global documentation on BioQ genomic features, such as the list of feature keywords and tables, can be found in web/perl/dbdoc/features.xml